Biology > STUDY GUIDE > Study Guide > University of California, San Diego - BENG 168beng 168 final notes. Chapter 2 – DNA, (All)
Study Guide > University of California, San Diego - BENG 168beng 168 final notes. Chapter 2 – DNA, RNA, and Protein Synthesis.
Document Content and Description Below
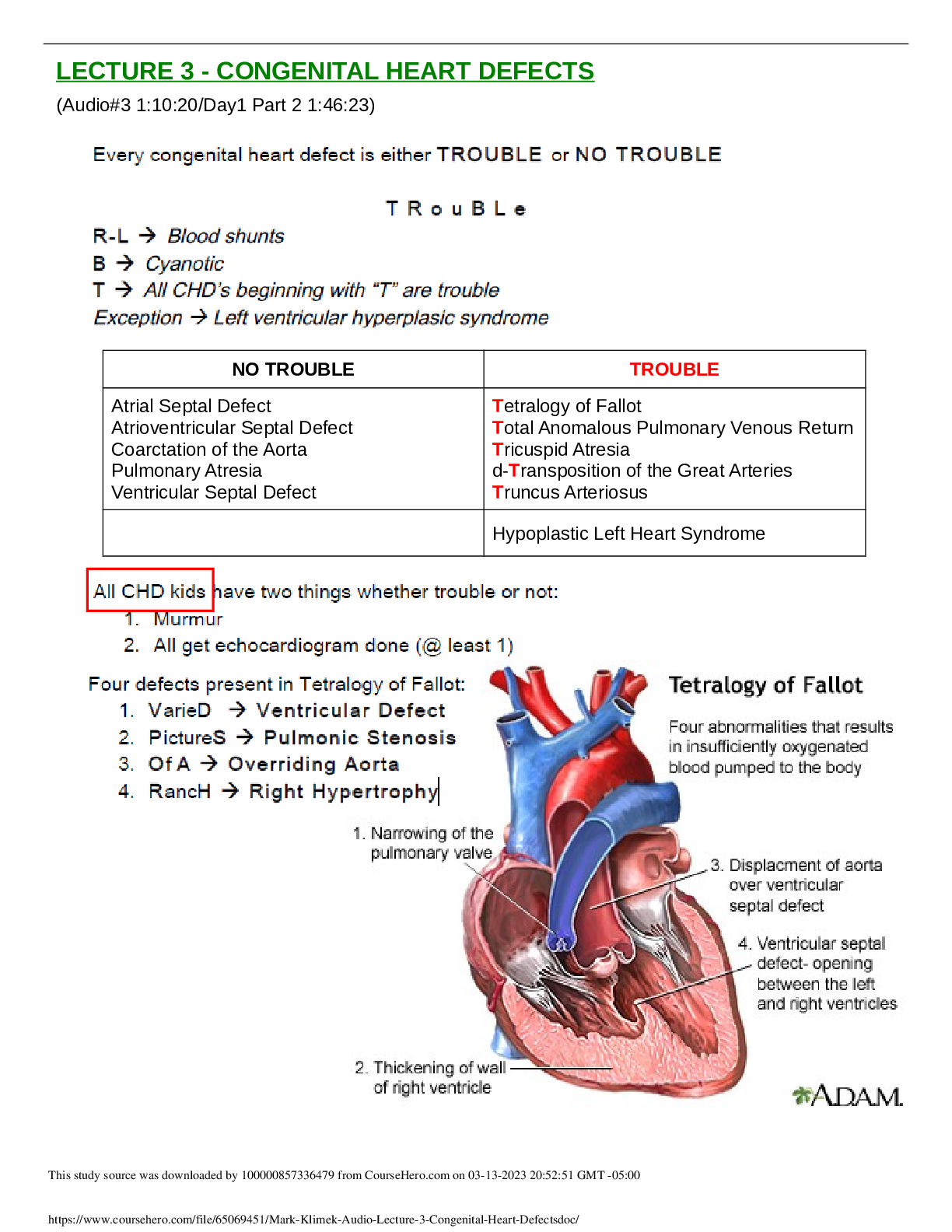
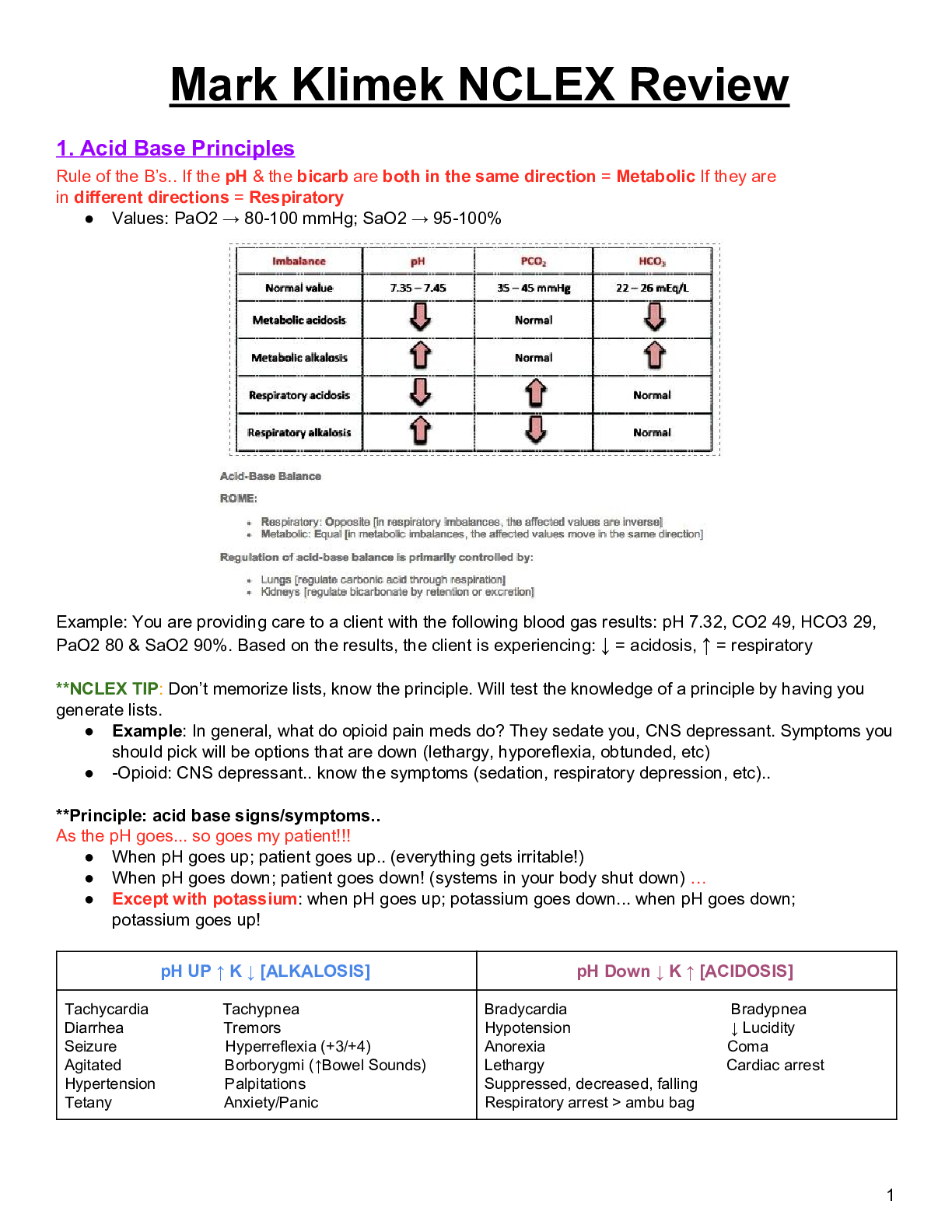
Chapter 2 – DNA, RNA, and Protein Synthesis • DNA structure o Composed of nucleotides – deoxyribose + phosphate + base o Bases – adenine (purine), guanine (purine), cytosine (pyrimidine), t... hymine (pyrimidine) A-T and G-C pairing 2 H bonds for A-T pair, 3 H bonds for G-C pair o Double-stranded helix (discovered by Watson + Crick using X-ray diffraction) • Replication o Each strand of existing DNA molecule is used as a template for producing a new strand o Sequence of growing strand is determined by base complementarity o Goes from 5’ to 3’ end on growing strand, new bases have to link up to 3’ OH on template strand o DNA polymerase binds nucleotides -> forms phosphodiester linkage o Prokaryotes vs eukaryotes: Prokaryotes have 1 ORI, eukaryotes have multiple ORI For eukaryotes, replication occurs in multiple sites simultaneously -> ends have to ligated together • Need telomerase to complete telomeres at ends of chromosomes • RNA, protein o Proteins – essential polymers, involved in almost all biological functions, consist of a specific sequence of amino acids Fold into specific conformations depending on locations of specific amino acid residues + overall AA composition Properties are determined by properties of side chains AA linked together by Peptidyl transferase o RNA – intermediate molecules for decoding genetic material, transcribed from DNA Have ribose instead of deoxyribose, uracil instead of thymine, only 1 stranded Types: mRNA, tRNA, rRNA • In prokaryotes, all transcribed by the same polymerase • In eukaryotes, each transcribed by different polymerases RNA polymerase links ribonucleotides together • Binds to promoter sequence to initiate transcription • Euk: alternative splicing gives rise to multiple proteins from same gene • Terminator sequence signals RNA poly to stop RNA synthesis Translation: • Prok: occurs simultaneously w/ transcription • Euk: occurs separately from transcription (transc in nucleus, transl in cytoplasm) • Primary transcripts in euk are modified w/ polyA tail and 5’ cap first • Reading of stop codon terminates translation -> termination factor recognizes stop codon and binds to ribosome Transcription regulation (bacteria) • Operon arrangement: contiguous bacteria structural genes that encode proteins required for several steps in a single metabolic pathway • Operator region: regulatory region that control on/off states of various operons, play essential role in determining if operon is transcribed • Effectors can bind to repressor proteins -> change conformation -> prevent it from binding to operator -> ”turn on” • Alternative: corepressor can bind to inactive repressor -> alter conformation to make it active -> turn off transcription Transcription regulation (eukaryotes) • Basal set of structural genes maintain routine cellular functions • Transcription mediated by proteins collectively classified as “transcription factors”, bind directly to DNA sequences • Promoters: TATA box, CCAAT sequence, GC box Chapter 3 – Recombinant DNA technology • Restriction endonucleases o Type II – restriction enzymes, bind to recognition sites n DNA and splice at these areas Can produce sticky ends (overhangs) or blunt ends IIS subcategory – cut a fixed number of nt away from the recognition site • Useful in preventing reopening of splice sites (nicks) Methylation of cytosine residues in host DNA prevents Res from cutting at sites Use ligase to catalyze formation of phosphodiester bonds at nicks in backbone • Plasmid cloning vectors o Plasmids – self-replicating, ds, circular DNA; maintained in bacteria as independent chromosomes Can be high-copy number or low copy number Plasmids from different incompatibility groups can be maintained in same cell Have the basic attributes to serve as vectors for carrying cloned DNA Have to be engineered first: • Insert unique RE sites for insertion of cloned DNA • Need selectable markers for identifying transformed cells o pBR322 resistance genes for Amp and Tet BamHI, HindIII, SaII, PstI, EcoRI RE sites E. coli ORI that functions only in E. coli, is maintained at a high copy number, cannot be readily transferred to other bacteria Process of cloning/transformation: • Insert GOI into Tet gene using RE site of choice o Final cells lack Tet resistance b/c gene is nonfunctional • Transform plasmid DNA into cells and plate on medium with Amp o Kills off cells w/o plasmid • Use replica plate on medium with Tet and identify colonies that die o Pick those colonies from original and culture o pUC19 Amp gene, lacZ gene, lac promoter, lacI gene, MCS site w/ many RE sites Cells w/o cloned plasmid should be able to hydrolyze X-Gal to blue color Nontransformed cells again die in presence of Amp Successfully transformed cells can’t hydrolyze X-Gal: lacZ gene is nonfunctional due to transformation disrupting codon sequence • Creating and screening a library o Treat a DNA sequence with RE and carry out partial digestions (low RE [] or short incubation time) o Clone the sequences into plasmid vectors o Identify clones w/ target sequences: DNA hybridization, immunological screening, assaying for protein activity, functional complementation • Cloning sequences encoding eukaryotic proteins: o Use reverse transcription to create cDNA for cloning (reverse transcriptase enzyme) o Secondary strands created from RNA-cDNA using ribonuclease to nick mRNA -> provide free 3’ OH group for DNA synthesis, 5’ activity of DNA polymerase I removes ribonucleotides that are encountered o Screen using DNA hybridization • Transformation methods o Electroporation – “electric-field mediated membrane permeabilization” o Conjugation – effective contact between donor cell and recipient cell, plasmid genes encoding conjugative functions Chapter 4 – Chemical Synthesis, Amplification, and Sequencing of DNA • Chemical synthesis of DNA o Phosphoramidite method 1) Linking first nucleotide to column -> 2) washing -> detritylation -> 3) washing -> 4) activation + coupling -> 5) washing -> 6) capping -> 7) oxidation -> n cycles of step 2 to step 7 -> step 2) -> remove oligo from colum -> purify o Uses of synthesized oligonucleotides Use as probes to screen genomic libraries Use for cloning DNA, creating unique cloning sites (linkers containing RE sites, adaptors for creating cloning sites in vectors) Key components for assembling synthetic genes • Uses: large scale protein production, testing protein function after altering codons, creating sequences w/ novel properties Creating full size genes • 1) Create set of overlapping, complementary nucleotides • 2) synthesize specified set of overlapping oligo, ~60 nt long, overlaps ~20 nt long • Polymerase Chain Reaction (PCR) o Amplification procedure for generating large quantities of DNA in vitro o Components: primers, template sequence, thermostable DNA polymerase, dNTPs o Steps Denaturation: 95 C, denature the template DNA Renaturation: 55 C, primers anneal to template strand Synthesis: 75 C, dNTPs start annealing to template strand using primers as starting point o Short copies eventually accumulate and outnumber longer copies o PCR amplification of full-length cDNAs Use oligonuc with oligo(dT) and added sequence (oligo(dT)-primer) + reverse transcriptase to create first cDNA strand Terminal transferase activity adds mostly dCs to end of each full-length first-strand cDNA molecule dG primer base pairs with dC tail, acts as template for extending first strand repeat cycles of forward and reverse primers o Gene synthesis by PCR Faster, more economical than using overlapping oligonucleotides w/ DNA polymerase + sealing nicks with ligase Start with 2 overlapping oligonucleotides, recessed 3’ OH groups serve as starting points for elongation -> annealing dNTPs Add two additional oligonucleotides that overlap with original 2 -> anneal dNTPs Can synthesize 1000 bp gene in one day • DNA sequencing techniques o Sanger: termination of growing strand using ddNTPs + fluorescence to detect where the strand stops elongating o Primer walking: have primers bind to sequence of interest and sequence the following segment Must sequence both strands for full accuracy False priming can lead to erroneous reading, use longer primers to avoid this Useful if you already have a general idea of the sequence o Pyrosequencing: NGS technique, basis is detection of pyrophosphate released during DNA synthesis Entails a series of enzymatic reactions, sequence 1 nt at a time Can be problematic for homopolymer tracts because number of nt is linear with amount of light produced -> can be difficult to gauge number accurately o Ligation: NGS, extend by ligation of short oligos in template dependent fashion Requires degenerate octamers or nonamers Each base type gives off a different fluorescence -> can do multiple bases at a time Perform multiple runs offset from each other by 1 nt • Large scale sequencing o Shotgun: fragment multiple copies of a DNA sequence with physical force -> end up with overlapping fragments Clone segments in vivo Use Sanger sequencing to sequence shorter fragments Use overlaps to reassemble full genome o Cyclic array (454 technology) Fragment genome into fragments, anneal to DNA capture beads Amplify using emPCR (PCR in emulsion bubble microreactors, more efficient) Place beads into wells and use pyrosequencing to sequence in parallel Chapter 5 – Bioinformatics, Genomics, Proteomics • GFP o Green fluorescent protein, can engineer to respond only in presence of substrate and use for operon/regulatory analysis • Functional genomics o DNA microarray technology Take cDNA from 2 or more samples, mix and hybridize to an array Each time has a different fluorescence attached to it Scan array for emissions and record Alternatively: screen cRNA synthesized from cDNA Purposes: • Identify genes whose expression changes in response to a particular biological condition • Identify genes that are coexpressed under different conditions/over a period of time o SAGE (serial analysis of gene expression) Use recombinant DNA techniques to clone randomly linked short sequences of cDNA to identify expressed genes • Proteomics o Separation and identification of proteins 2D PAGE: gel electrophoresis for separating proteins (by MW and charge) Use Mass spectrometry to identify proteins after separation • Measure mass/charge ratio, search for matches in protein database o Protein expression profiling: 2d differential in-gel electrophoresis • Similar to PAGE, but compare results from two samples ICAT method • Use affinity tag and labeled linker (light or heavy labeling) • Abundance of light/heavy ICAT indicates relative abundance of source proteins [Show More]
Last updated: 2 years ago
Preview 1 out of 35 pages

Buy this document to get the full access instantly
Instant Download Access after purchase
Buy NowInstant download
We Accept:

Reviews( 0 )
$13.00
Can't find what you want? Try our AI powered Search
Document information
Connected school, study & course
About the document
Uploaded On
Feb 22, 2021
Number of pages
35
Written in
Additional information
This document has been written for:
Uploaded
Feb 22, 2021
Downloads
0
Views
119







.png)











