Biology > Lab Report > Genetics Lab Report II (Biol 203, Spring LATEST UPDATE) PCR Cloning and Expression of a GFP fusion p (All)
Genetics Lab Report II (Biol 203, Spring LATEST UPDATE) PCR Cloning and Expression of a GFP fusion protein in E. coli
Document Content and Description Below
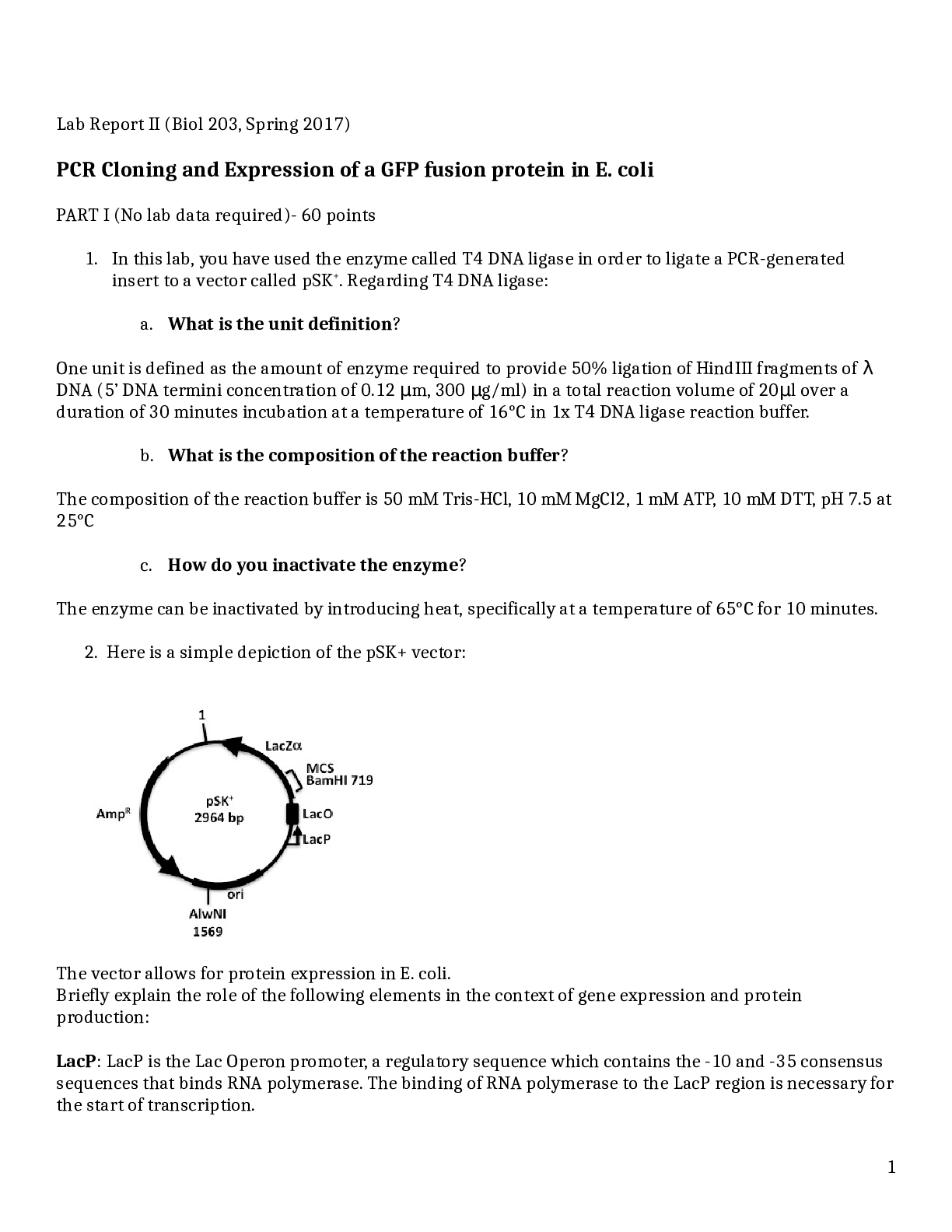
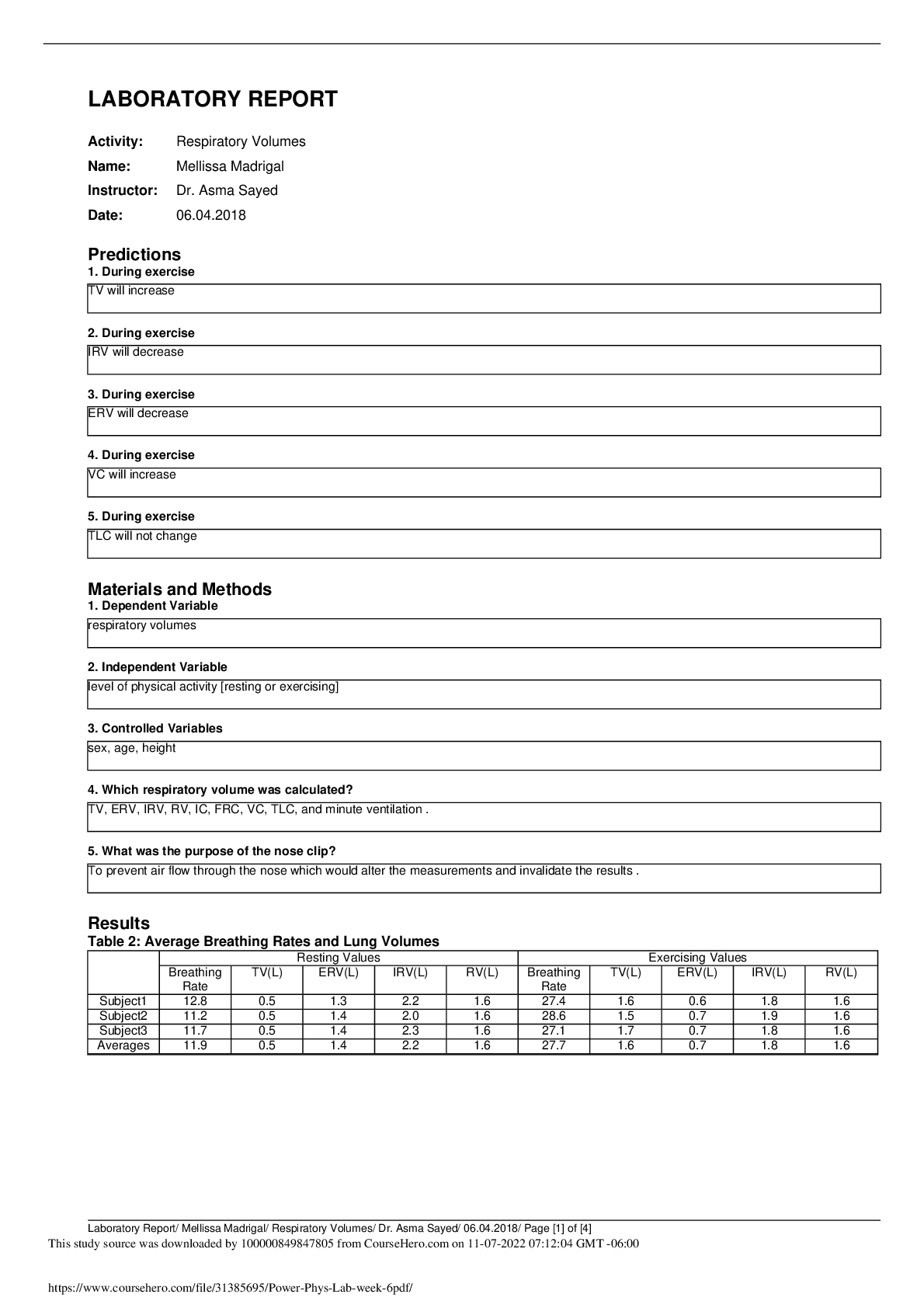
Lab Report II (Biol 203, Spring 2017) PCR Cloning and Expression of a GFP fusion protein in E. coli PART I (No lab data required)- 60 points 1. In this lab, you have used the enzyme called T4 DNA l ... igase in order to ligate a PCR-generated insert to a vector called pSK+. Regarding T4 DNA ligase: a. What is the unit definition? One unit is defined as the amount of enzyme required to provide 50% ligation of HindIII fragments of λ DNA (5’ DNA termini concentration of 0.12 m, 300 g/ml) μ μ in a total reaction volume of 20 l over a μ duration of 30 minutes incubation at a temperature of 16°C in 1x T4 DNA ligase reaction buffer. b. What is the composition of the reaction buffer? The composition of the reaction buffer is 50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, pH 7.5 at 25°C c. How do you inactivate the enzyme? The enzyme can be inactivated by introducing heat, specifically at a temperature of 65°C for 10 minutes. 2. Here is a simple depiction of the pSK+ vector: The vector allows for protein expression in E. coli. Briefly explain the role of the following elements in the context of gene expression and protein production: LacP: LacP is the Lac Operon promoter, a regulatory sequence which contains the -10 and -35 consensus sequences that binds RNA polymerase. The binding of RNA polymerase to the LacP region is necessary for the start of transcription. 1LacO: LacO is the Lac operator region, a regulatory sequence that the repressor protein binds. When the repressor protein is bound to LacO, no transcription takes place because RNA polymerase cannot bind to the LacP region. The operator region overlaps with the promoter region, thereby influencing the binding of RNA polymerase. LacZ: In terms of gene expression, LacZis responsible for determining whether or not a protein was inserted in the proper orientation. If a protein is in the correct orientation in respect to LacZ then there α is a translational fusion, however if it is not in the correct orientation then no fusion protein is produced. LacZ is a gene on the lac operon that is responsible for the production -Galactosidase, which breaks β down lactose into glucose and galactose. Briefly explain and indicate what is required for high expression in E. coli in this context. For high expression of E. coli it is necessary to have an abundance of lactose opposed to glucose via the growth medium to which the E. coli is introduced. In high levels of lactose and low levels of glucose, lactose will transform into allolactose, which binds to the repressor protein and thereby inactivates it. When lactose is present, allolactose or IPTG act as the inducers of the expression of the lac genes because they bind to the repressor and remove it, allowing transcription to proceed. The repressor protein is then unable to bind to LacO which allows RNA polymerase (which is now free) to carry out transcription. When there is no lactose present in the cell (or high levels of glucose), the repressor protein is bound to the operator and will not allow RNA polymerase to transcribe the lac genes. In order to increase rate of transcription, the CAPP-cAMP complex is used. The vector also contains other important elements. Explain their role: AmpR: A gene that promotes ampicillin resistance, thereby allowing growth on Ampicillin plates. ori: Represents the origin of replication which allows DNA replication in bacteria. 3. You plated your transformations on agar containing Ampicillin, IPTG and X-Gal. -Explain the use of IPTG: IPTG is a structural analog of allolactose and acts as an inducer, therefore it is used to bind to the lac repressor and allow lac transcription. -Explain the use of X-Gal: 2X-Gal is utilized in the medium due to its ability to turn blue when bacteria has a functional LacZ gene and is producing -Galactosidase. β 4. You are attempting to clone an EcoRI fragment into an EcoRI-restricted vector treated with CIP, and you obtain the following results on your transformation plates (with Amp): #1 vector only 107 colonies +ligase #2 vector+insert+ligase 663 colonies - Would you expect more, fewer or about the same number of colonies if no ligase were added in #2? Explain. If no ligase was added to #2 then there would be fewer colonies. The vector and insert contain complementary ends, therefore without the presence of ligase the free 5’ phosphate of the vector would not join with the 3’ hydroxyl end of the insert. - Would you expect more, fewer or about the same number of colonies with #1 with a vector not treated with CIP? Explain. An untreated vector + ligase would result in more colonies than present in #1. This is because without a CIP treated vector the DNA is free to recirculate during ligation. 5. You wish to clone an insert (“insert “X”) by PCR-cloning, into the pSK+ vector. The Start and Stop codons for the open reading frame (1540 bp) you want to clone as insert X are indicated by a green circle and a Stop sign, respectively. Below is a restriction map for the region, showing the sites for the indicated restriction enzymes: HindIII EcoRV XhoI XhoI SpeI XbaI KpnI HindIII : approx.. 100 bp No restriction sites: [Show More]
Last updated: 3 years ago
Preview 1 out of 15 pages
Buy this document to get the full access instantly
Instant Download Access after purchase
Buy NowInstant download
We Accept:

Reviews( 0 )
$7.00
Can't find what you want? Try our AI powered Search
Document information
Connected school, study & course
About the document
Uploaded On
Apr 24, 2021
Number of pages
15
Written in
All
Additional information
This document has been written for:
Uploaded
Apr 24, 2021
Downloads
0
Views
87

.png)










.png)





.png)



