Biology > STUDY GUIDE > BIOL 3362 STUDY GUIDE TEST 1 (Biochemistry, Membranes, Plants, Bacteria, Calvin Cycle, Chloroplast, (All)
BIOL 3362 STUDY GUIDE TEST 1 (Biochemistry, Membranes, Plants, Bacteria, Calvin Cycle, Chloroplast, Photosynthesis, Amino Acids).
Document Content and Description Below
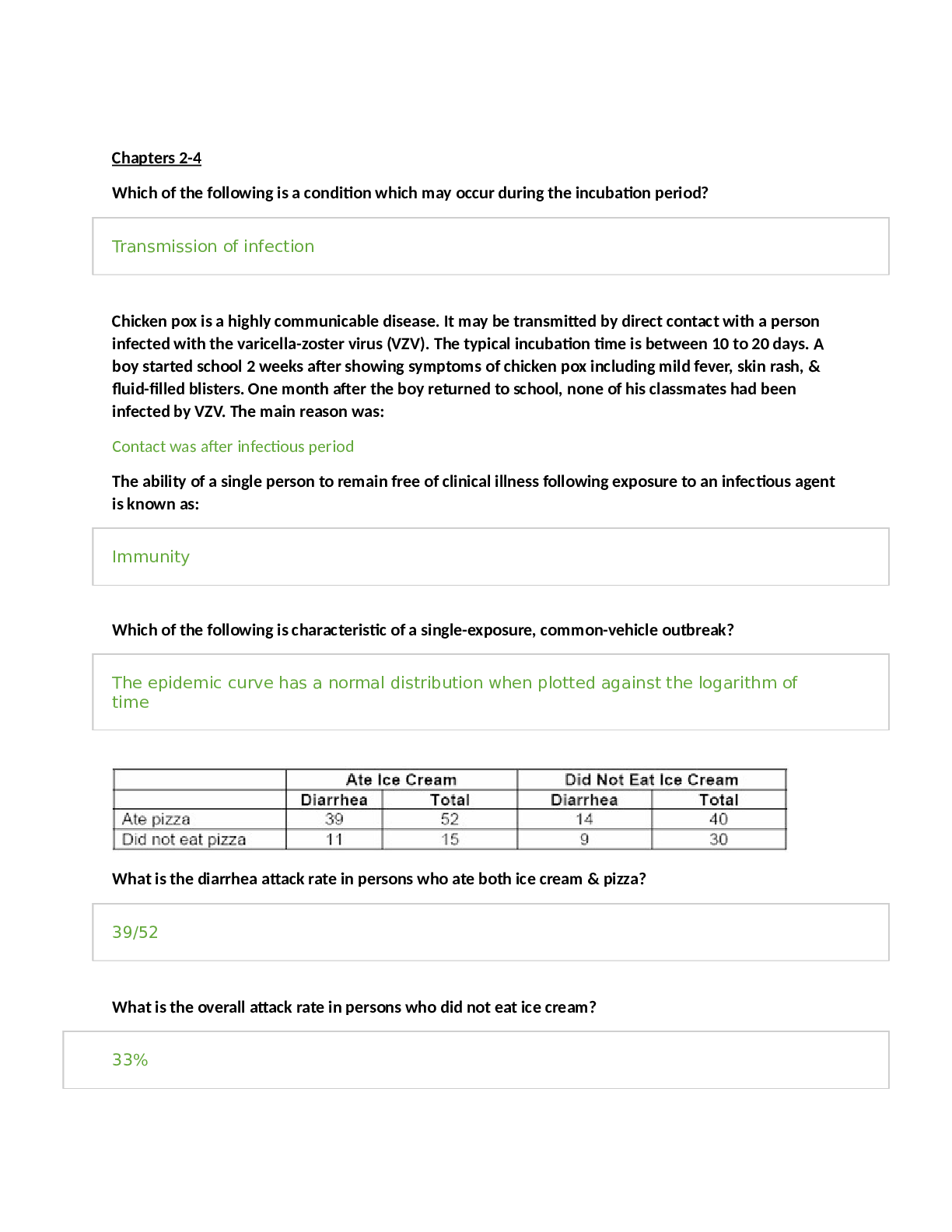
STUDY GUIDE TEST 1 CHAPTER 21: PHOTOSYNTHESIS Where does photosynthesis occurs in plants and bacteria? • Plants- chloroplasts o Light reactions in the thylakoid membrane o Dark reactions in the... stroma • Bacteria- o Cyanobacteria- thylakoid membranes in cytoplasm o Photosynthetic bacteria- in membranes in the cytoplasm Chloroplasts structure and function are very important. Inner membranes are important for photosynthesis to occur because of the separation of charges. Outer membrane - It is a semi-porous membrane and is permeable to small molecules and ions, which diffuses easily. The outer membrane is not permeable to larger proteins. Intermembrane Space - It is usually a thin intermembrane space about 10-20 nanometers and it is present between the outer and the inner membrane of the chloroplast. Inner membrane - The inner membrane of the chloroplast forms a border to the stroma. It regulates passage of materials in and out of the chloroplast. In addition of regulation activity, the fatty acids, lipids and carotenoids are synthesized in the inner chloroplast membrane. Stroma is an alkaline, aqueous fluid which is protein rich and is present within the inner membrane of the chloroplast. The space outside the thylakoid space is called the stroma. The chloroplast DNA chloroplast ribosomes and the thylakoid system, starch granules and many proteins are found floating around the stroma. Thylakoid System The thylakoid system is suspended in the stroma. The thylakoid system is a collection of membranous sacks called thylakoids. The chlorophyll is found in the thylakoids and is the sight for the process of light reactions of photosynthesis to happen. The thylakoids are arranged in stacks known as grana. Each granum contains around 10-20 thylakoids. What are light and dark reactions? Light reactions (without CO2) are the redox reactions which produce oxygen, ATP and NADPH using a proton gradient and electron transport chain (in thylakoid membrane) (water is electron source for the production of ATP and NADPH). The ATP and NADPH are then used in the dark reactions to fix CO2 and produce hexose. (dark reactions occur in stroma) What are the products of photosynthesis and how are they made? What is the source of electrons? Oxygen and hexose. Oxygen is made during the photolysis of water, and hexose is made during the dark reactions of photosynthesis. H2O is the electron source In bacteria, the electron sources can come from : H2S , isopropanol, or other oxidizable compounds What is chlorophyll? Chlorophyll is a pigment found within chloroplasts that harvests light and funnels electrons to a reaction center via resonance energy transfer to a reaction center where they are used for chemical synthesis. What is chlorophyll function? To use its delocalized pi electrons above and below the ring structure of chlorophyll to absorb photons of light and transfer excited electrons to a reaction center where they can be used for chemical synthesis What does chlorophyll contain (in terms of metals, and functional groups)? It is a substituted tetrapyrrole ring system coordinated with a magnesium atom. (Correction : Modified tetrapyrolle ring w/ hydrophobic tail/5 rings w/ Mg) The hydrophobic aliphatic region/tail anchors it What are fates of excited electrons? (4 fates) 1. Heat loss - e- goes to lower state & increases vibrational energy 2. Loss as light / Fluorescent light - longer/less energetic - occurs only in saturating light 3. Resonance energy transfer - excitation of e- neighboring molecule - permits energy transfer to one reactive site and exciton transferred 4. Energy transduction - more energetic molecule = better e- donor (electron is turned into a reducing agent) / transfer of light energy to chemical energy What is a reactive center? Contains 2 photochemically reactive chlorophyll a molecules. Chemical synthesis occurs here. What are the two photosystems and what are their functions? What is the Z scheme? PSI - is a very strong reducing agent (e- donor) that reduces NADP to NADPH PSII - is a very strong oxidizing agent (e- acceptor) that splits H2O, which produces O2. PSII transfers e- to PSI. This process pumps protons via the electron transport chain, which can be used for ATP synthesis. Z-Scheme - electron transfer follows a Z scheme (up-down-up) with respect to reduction potentials. The up changes require energy input. (Understand this process in detail) More positive reduction potential = more likely to be reduced. What are the similarities between electron transport in oxidative phosphorylation and photosynthesis? What components are in common? • Both use a proton gradient to generate ATP • Flow of electrons through redox intermediates, membrane bound carriers (quinones), cytochromes and Fe-S proteins Ferredoxin shows up a lot. Why? It’s the Fe-S protein that mediates electron transfer What is the difference between cyclic and noncyclic photophosphorylation? Cyclic photophosphorylation does not evolve oxygen or produce NADPH. It only shuttles electrons between photosystem one and the cytochrome b6f complex. It only provides the proton gradient by which ATP is synthesized. Non cyclic photophosphorylation produces both oxygen and NADPH. Cyclic photophosphorylation is 20 times slower than noncyclic photophosphorylation but may overcome the ATP deficit of the Z scheme. Also, it has an additional protein, cytochrome b6. Where are protons pumped in the process? What steps, and into what compartments? Protons are pumped from the stroma into the lumen of the thylakoid. They are pumped when plastoquinone delivers electrons to the Cytochrome b6f complex (4 protons). The energy from the redox reactions performed is used to drive H+ pumping. Protons are also deposited into the lumen following the photolysis of water. Carbon dioxide fixation. Which reactions incorporate CO2? What are the unique reactions of the Calvin-Benson cycle? Ribulose-1,5-bisphosphate is used to accept CO2 and form 2 3-phosphoglycerates. This reaction is catalyzed by rubisco. The unique reactions of the Calvin cycle are: • Rubisco • P-glycerate kinase(glycolytic enzyme) o Consumes 12 ATP • NADPH-dependant GAP dehydrogenase o Accounts for all NADPH consumed by Calvin cycle • Carbon rearrangements o Done by transketolase + aldolase = works in reverse of glycolysis o Produces 7-carbon intermediate o The acting phosphatase is unique to plants • Ribulose-5-p kinase / phosphoribulose kinase o Produces RuBP o 6 ATP Where are ATP and NADPH used? P-glycerate kinase (actually a glycolytic enzyme). It consumes 12 ATPs. NADPH-dependent GAP dehydrogenase (note that a glycolytic enzyme uses NAD and catalyzes the reverse reaction). This accounts for all the NADPH consumed by the Calvin cycle. The carbon rearrangements are mediated by transketolase and aldolase, which works in reverse of glycolysis and can generate a 7-carbon intermediate. A phosphatase acting on S-1,7- bisP is unique to plants. Finally, ribulose-5-P kinase to make RuBP, which accounts for 6 ATPs. 12 ATP are used in the conversion of 3-phosphoglycerate to 1,3-bisphosphoglycerate and 6 ATP are used when regenerating ribulose-1,5-bisphosphate from ribulose-5-phosphate.12 NADPH is used when converting 1,3-bisphosphoglycerate to glyceraldehyde-3-phosphate. What are the reactions of Rubisco? How is Rubisco activity controlled? Rubisco can react with oxygen or CO2. Reaction with CO2 yields 2 3-phosphoglycerates. Reaction with oxygen yields 3-phosphoglycerate and glycolate. Rubisco is regulated by light, RuBP, CO2, and rubisco activase. RuBP binds tightly to the non-carbamylated form of rubisco and inhibits it. Rubisco activase removes RuBP and activates rubisco. Light activates rubisco activase, and thus, Rubisco. How does light control CO2 fixation? Thioredoxin and ferrodoxin involvement is this control. Light Increases pH in the stroma, which activates many enzymes in the stroma Light generates reducing power in the form of reduced ferrodoxin, which leads to reduced thioredoxin, which, in turn, activates Calvin cycle enzymes Light induces movement of Mg2+ into the stroma and activates key Calvin cycle enzymes (Rubisco and FBPase) How do plants avoid the problem of the reaction of Rubisco with oxygen? Through use of a C-4 intermediate instead of a C-3 intermediate. CO2 combines with PEP to form Oxaloacetate. OAA is then converted to Aspartate or Malate. Malate is decarboxylated, and the liberated CO2 is used in the Calvin Cycle in the bundle sheath cells, far away from the oxygen rich surface. How do bacteria avoid the problem of the reaction of Rubisco with oxygen? (see lecture notes, since this is not in the book) Through use of CO2 concentrating mechanisms called carboxysomes. CO2 is converted to HCO3- upon entering the cell. Once HCO3- enters the carboxysome, it is converted by carbonic anhydrase to CO2. This allows for maximum CO2 influx and minimal O2 influx. Also know the location of the various photosynthesis components. New slide (24) is important: Lateral heterogeneity in thylakoid membrane organization (A) Electron micrograph of the thylakoid membrane showing stacked grana and unstacked stromal lamellae regions. (B) Model showing the distribution of the major complexes of photosynthetic electron and proton transfer between the stacked grana and unstacked stromal lamellae regions. CHAPTER 25: Nitrogen acquisition The nitrogen cycle: • Nitrate Assimilation: the conversion of nitrate to ammonia. It occurs in plants, some fungi, and some bacteria. o How many electrons involved and what metals are involved? What are the source of electrons? ▪ The first step is a two-electron reduction of nitrate to nitrite, catalyzed by nitrate reductase, from NADH to -SH to FAD to cyto b557 to MoCo and finally to nitrate. ▪ The second step is a six-electron reduction of nitrite to ammonia, catalyzed by nitrite reductase, from photosynthetically reduced ferredoxin to 4Fe-4S cluster to siroheme and finally to nitrite. • Nitrogen Fixation: the conversion of N2 to ammonia. It occurs only in bacteria. Sometimes these bacteria associate with plants. This is an anaerobic process. o What organisms do this? ▪ It is only done by bacterial cells. o What are its requirements (reductant, energy, etc)? ▪ All nitrogenases require the enzyme nitrogenase, a strong reductant (such as reduced ferrodoxin), ATP, and O2-free conditions. ▪ Depending on the bacterium, electrons for N2 reduction may come from light, NADH, hydrogen gas, or pyruvate. o What are its components (the two protein components), and what do they do? What factors control and affect its activity? ▪ The Fe-protein (nitrogenase reductase hydrolyzes two ATP per electron transferred, or 16 per N2. The ATP is used for overcoming the activation energy required to break the N2 bonds.This component has a single 4Fe- 4S cluster, and is very O2 sensitive.) and the MoFe-protein (nitrogenase is an a2b2 protein. Each ab dimer contains a P-cluster and the FeMo- cofactor. This component is also O2 sensitive.) ▪ Two factors commonly control nitrogenase: ▪ ADP inhibits activity. ▪ NH4+ inactivates a transcriptional activator. ▪ Some organisms have a third mechanism, ADP-ribosylation of the nitrogenase reductase subunit. • Nitrification: Bacteria then can take the reduced nitrogen, as ammonia, and oxidize it. Nitrifying bacteria convert ammonia to nitrite and nitrate, and use the energy released for growth. This process is called nitrification because it makes the nitrogen more readily available to plants. Nitrate is more soluble than ammonia, and plants absorb it more quickly. • Denitrification: the conversion of nitrate to N2, which is released to the atmosphere. It is done by denitrifying bacteria, which are very efficient. What pathways assimilate ammonia and what are the products? What are the differences between the two pathways? What is the reductant? Know the reaction mechanism of glutamine synthetase since is is found for many enzymes. The primary differences are that one requires more energy than the other, and the former can assimilate a low concentration of ammonia. The GDH pathway synthesizes glutamate and glutamine by the actions of GDH and GS. GDH has a high Km for ammonia, whereas GS has a much lower Km. With high ammonia (not normal in nature), GDH can assimilate ammonia into glutamate, and GS can assimilate ammonia into glutamine. The GS-glutamate synthase pathway: When ammonia is low, which is normal in nature, this pathway first synthesizes glutamine via GS from ammonia and glutamate. Then, glutamate synthase, uses glutamine as a nitrogen donor for the formation of glutamate. The difference in this pathway is energy consumption. The GDH pathway does not need to consume ATP for glutamate synthesis. The GS-glutamate synthase pathway does. It can be calculated for E. coli that the difference in energy is 10% of the cell’s total energy requirement. What are the intracellular nitrogen donors, and their relative quantitative importance. With high ammonia, the cell needs 75% of its nitrogen as glutamate, and 25% as glutamine. Therefore, the GDH pathway assimilates 75% of the ammonia, and GS 25%. With low ammonia, the GS-glutamate synthase pathway and GS assimilates 100% of the ammonia, and converts 75% of the glutamine made to glutamate. How is glutamine synthetase controlled? (I will not ask specific inhibitors, but what properties are shared by the inhibitors.) What is the effect of high and low glutamine on these controls? • Principles of feedback inhibition o Gly and Ser will accumulate if the cell has enough purines, which require glutamine for their synthesis. Why alanine inhibits is not known. These amino acids bind to the glutamate site. The other compounds require glutamine for their synthesis, and are endproducts. • The covalent modification o Adenylylation inactivates GS. Glutamine allosterically controls a bifunctional regulatory enzyme, uridylyltransferase (UTase)/uridylyl-removing enzyme (UR) which controls the activity of another regulatory protein, PII, which in turn controls ATase activity. With high glutamine, UR is active and results in the adenylylation of GS. With low glutamine, UTase is active and results in deadenylylation of GS. Glutamine is the major effector. • Gene expression controls: what metabolites are involved? o High glutamine (nitrogen excess) results that the gene for GS is not expressed. Low glutamine (nitrogen limitation) has the opposite effect. NRI~P is a transcriptional activator that turns on the gene for GS. It is the level of glutamine that is important, not the ratio of glutamine to a-ketoglutarate. Transaminations: • Nitrogen donor: addition of a nitrogen from glutamate to an a-keto acid by transamination • Cofactor: The aminotransferases require pyridoxal phosphate as an essential cofactor. • Reversibility: The reactions are usually readily reversible. Glutamate is virtually always in excess, which determines the direction of the reaction. Amino acid classifications: families, and members of each family • Alpha-Ketoglutarate family: glutamate, glutamine, proline, arginine, lysine • Oxaloacetate family (Aspartate family): Aspartate, asparagine, methionine, threonine, isoleucine, lysine • Pyruvate Family: Alanine, Valine, Leucine, Isoleucine • 3-Phosphoglycerate family: Serine, Glycine, Cysteine • Phosphoenolpyruvate & Erythrose-4-P Family: (the aromatic amino acids) Phenylalanine, Tyrosine, Tryptophan • Histidine family: histidine Essential and nonessential amino acids (what does this mean), difference between plants, bacteria and higher animals. Do not memorize the amino acids for this classification. Mammals can synthesize only about 10 of the amino acids. These are called the nonessential amino acids. The ones that can’t be synthesized are essential, and mammals lack the pathways. Plants can synthesize all 20 amino acids. Similarities in the pathways of arginine and proline synthesis, cyclization and its prevention, mechanism of activation, biological reductant. Y-carboxyl group activated by phosphorylation Activated carboxyl reduced to aldehyde (requires NADPH) Aldehyde interacts with a-amino group and cyclizes (note: for arginine.. Orinthine is synthesized from glutamate… N-acylation blocks cyclization in proline synthesis then steps are similar to proline synthesis) Carbamoyl-P synthetase, precursor for which compounds, most unusual aspect of reaction. How many enzymes in higher animals, and where are they located. It consumes two ATPs which is unusual. CPS-I is a mitochondrial enzyme in higher organisms, probably because this is where the ATP is. It uses ammonia as a nitrogen source. E. Coli has only one CPS, and it uses glutamine as a N source. The carbon source is bicarbonate. Final steps in arginine synthesis: common reactions with urea cycle. Addition of urea group to N end of ornithine Urea cycle: • which steps in mitochondria, and its function? o Carbomylation - NH3+ group of Ornithine is carbomylated by a carbomyl group from Carbamoyl-P synthesized by CPS-1 in the mitochondria • how nitrogen enters the cycle? • Ammonia, glutamate and aspartate • what tissue has enzymes of the cycle? • Liver Aspartate family: • similarities with glutamate (α-ketoglutarate family) - almost all chemistry is at the terminal carboxyl group and requires activation of the carboxyl group by similar mechanisms An Amino group is transferred from glutamate to oxaloacetate via PLP- dependent aminotransferase rxn • asparagine synthesis (mechanism of activation) - synthesized similar to GS reaction Carboxyl group is activated with ATP forming aspartyadenylate o Amidation of B-carboxyl group of aspartate • control of first reaction of common pathway o Each of the 3 aspartokinases are inhibited by different end products o Allows for continued synthesis of other amino acid intermediates o Methionine synthesis, how sulfur is added, vitamins and cofactors involved, mechanisms that result in accumulation of homocysteine, methyl group addition, transsulfuration pathway. • OH activated by succinylation with succinyl-CoA • cysteine (universal S donor in bacteria) replaces succinyl-CoA • cysteine removed (-SH remains) --- This produces homocysteine • Homocysteine -SH is methylated by N5-methyl-THF as the methyl donor to produce methionine Pyruvate family: • alanine synthesis o Glutamate-Alanine transaminase o Transamination of pyruvate to Alanine is readily reversible • precursors for branched chain amino acids o Hydroxyethyl TPP and either Pyruvate or alpha-ketobutyrate • role of thiamine pyrophosphate and of shared enzymes • leucine synthesis pathway mimics what pathway. • TCA cycle 3- phosphoglycerate family: • serine, glycine, and cysteine synthesis o All derived from 3-phosphoglycerate (created by 3PG Dehydrogenase) o Serine: ▪ 3PG is oxidized by NAD+, then transaminated by glutamate to generate 3-phosphoserine ▪ Serine phosphatase generates serine ▪ Serine inhibits 3PG Dehydrogenase o Glycine (2 ways to synthesize Glycine) ▪ Serine’s B-carbon is transferred to THF generating Glycine and N5,N10 -methylene THF ▪ Glycine can be synthesized by reversal of Glycine oxidase rxn (N5,N10-methylene-THF + NH4+ + CO2) o Cysteine : ▪ Sufhydryl transfer to erine ▪ Bactera can use H2S • synthesis of C1 intermediates o • sulfur assimilation (what compound is environmental sulfur source, what is actually incorporated into amino acids). o Sulfate-environmental source o Sulfide-amino acids • What is the major donor for sulfur in various biosynthesis? Cysteine Aromatic family: • Precursors o Erythrose-4-P and PEP • end product of common pathway o Chorismate • regulation of first reaction, (can ignore tryptophan synthesis, except know which compounds are required for it synthesis) o First reaction inhibited by amino acid end products o Tryptophan precursors-PRPP and serine • conversion of phenylalanine and tyrosine o Occurs by phenylalanine monooxygenase • disease that results from problems with the interconversion o Phenylketonuria Histidine synthesis: • Precursors o PRPP and ATP • role of ATP o Provides one carbon and nitrogen The logic of herbicides (what type of pathways targeted), but knowledge of specific herbicides is not required. Target pathways found only in plants but not humans so that pathways affected only target plants. Amino acid degradation: • what is difference between glycogenic and ketogenic amino acids o Glycogenic amino acids can be converted to glucose, while ketogenic acids are converted to ketone bodies or fatty acids upon degradation • how nitrogen is removed, and where does it go? o Transamination ▪ Makes glutamate-2 paths it can go from here ▪ Oxidatively deaminated by glutamate dehydrogenase (cofactor: NAD) → makes alphaKG+ammonia, ammonia enters urea cycle as carbamoyl P ▪ Donate the N to Oxaloacetate → makes aspartate (provides N for argnine synthesis in urea cycle), aspartate forms fumarate-->back to oxaloacetate • alkaptonuria and how are symptoms prevented? o Amino acid synthesis disease o When 1 enzyme of tyrosine metabolism is blocked=dark cartilage tissue, black urine o Prevention: intake of phenylalanine=increase homogenoic acid Structures to be able to recognize first intermediate in pathways of: • synthesis of proline (glutamyl-P) • arginine (N-acetylglutamate) • asparagine (aspartate) • lysine-threonine-methionine common pathway (aspartyl-P) • methionine (homoserine) • alanine (pyruvate) • serine (3-phosphohydroxypyruvate) • cysteine (O-acetylserine) • glycine (serine) • histidine (N1-5’-phosphoribosyl-ATP) Structures to be able to recognize last intermediate in synthesis of: • proline (D1-pyrroline-1-carboxylate) • arginine (argininosuccinate) • methionine (homocysteine) • serine (3-phosphoserine) • phenylalanine (phenylpyruvate) • tyrosine (4-hydroxyphenylpyruvate) • tryptophan (indole) • histidine (histidinol) [Show More]
Last updated: 2 years ago
Preview 1 out of 13 pages
.png)
Buy this document to get the full access instantly
Instant Download Access after purchase
Buy NowInstant download
We Accept:

Also available in bundle (1)

BIOL 3362 - Exam 1 -4 Study Guide BUNDLE | Download To Score A.
BIOL 3362 - Exam 1 -4 Study Guide BUNDLE | Download To Score A.
By Goodluck Academia 3 years ago
$14
5
Reviews( 0 )
$12.50
Can't find what you want? Try our AI powered Search
Document information
Connected school, study & course
About the document
Uploaded On
Sep 22, 2021
Number of pages
13
Written in
Additional information
This document has been written for:
Uploaded
Sep 22, 2021
Downloads
0
Views
85









.png)

.png)


.png)






